The microorganisms living in, on, and around us, collectively known as the microbiome, produce metabolites that affect our health. Some of these molecules are essential for our wellbeing, while others can harm us. Colibactin is an example of a toxin, produced as part of microbial warfare, which induces double-stranded breaks in the DNA of cells in the intestinal epithelium and is correlated with higher incidences of diseases such as colorectal cancer.
Bacteria producing colibactin must protect their own DNA, so they assemble a non-toxic precursor (precolibactin) in their cytoplasm and convert it to the active toxin only after exporting it outside the cell. Central to this resistance is the prodrug motif, a chemical group installed by the first enzyme of the colibactin biosynthetic pathway and removed by the ClbP peptidase (so named because it is gene P in the colibactin biosynthetic gene cluster) in the periplasmic space. The prodrug motif is like the safety pin of a grenade: while it is in place precolibactin is innocuous, but its removal by ClbP triggers a chemical reaction that forms the DNA-alkylating groups (warheads) in colibactin. This prodrug resistance mechanism is also used in the biosynthesis of other toxins by a variety of bacteria.
A collaboration between the Gaudet (MCB department) and Balskus (CCB department) groups, awarded the Björkman-Strominger-Wiley prize last year, aims to understand how ClbP and related enzymes activate their respective toxins and the effects of inhibiting them on toxin activation and on the complex microbial communities where these toxins mediate competition between organisms. In a pair of Nature Chemical Biology papers, the groups report structures and mutagenesis analysis of ClbP which reveal how it functions (Velilla et al.) and a set of novel molecules that mimic the prodrug motif and selectively inhibit ClbP and related prodrug-activating peptidases (Volpe et al.).
Determining the 3D structure of membrane proteins like ClbP is notoriously challenging. Previous studies tried to side-step the issue, focusing on its isolated soluble domain. While useful, these structures did not explain why the transmembrane domain of ClbP is indispensable to its enzymatic function. The full-length crystal structures of ClbP reported in Velilla et al. show how its transmembrane domain interacts with the prodrug motif, demonstrating its essential role in substrate binding. The structures also reveal an intricate hydrogen-bonding network in the ClbP active site that is responsible for its exquisite specificity towards the d-asparagine amino acid present in its prodrug motif. To provide further proof of this specificity mechanism, Velilla et al. showed that changes to this network of interactions, which is strictly conserved in peptidases that activate d-asparagine prodrug toxins, reprograms the specificity of ClbP.
The authors discovered that ClbP forms a homodimer in which the two subunits face each other and form a dome-shaped cavity that is flanked by the catalytic sites of each subunit (Figure). While the crystal structure provided the first clue, a cryogenic electron microscopy (cryo-EM) structure of ClbP provided definitive proof that ClbP is a dimer. This discovery was especially exciting since chemists from the Balskus group and the Crawford group at Yale recently proposed that ClbP’s substrate, precolibactin, is itself a pseudodimer containing two prodrug motifs that is activated into a double-warhead colibactin. The chemical structure of colibactin is a puzzle which has taken over a decade to assemble and which has drawn information from the fields of bioinformatics, enzymology, and total synthesis. The discovery of ClbP dimerization further supports the proposed structure of precolibactin and raises new questions about the regulation of colibactin activation and whether the dimerization of ClbP is relevant to this process.
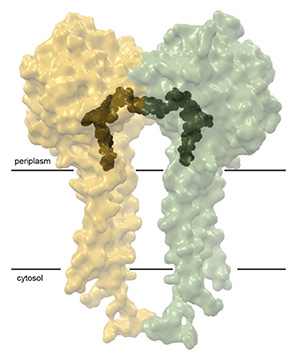
Transparent surface representation of the ClbP dimer, with one subunit in yellow, and the other in green. A model of the precolibactin substrate is docked in the two active sites (black). The black bars illustrate the approximate position of the cellular membrane.
Because ClbP performs the last step in the biosynthesis of colibactin, blocking its activity has been the go-to approach in metabolomics studies aiming to determine the structure and biological activity of colibactin. Historically, this has been achieved by deleting the clbP gene. This genetic approach is quite blunt and is not amenable to studying colibactin in the context of its native producers or its effect in the composition of complex microbial communities. The Volpe et al. paper describes the development of boronic acid mimics of the prodrug motif which potently inhibit ClbP by forming covalent adducts with its catalytic serine. A crystal structure by the Gaudet group demonstrates the covalent adduct.
The inhibitors block colibactin production and prevent genotoxicity in eukaryotic cells. They are non-toxic and selective towards ClbP and related peptidases, which makes them ideal to study complex microbial communities and highlights their potential use for preventing diseases associated with colibactin production in the gut. Because the inhibitors are recognized by amino acid residues that are strictly conserved in other prodrug-activating peptidases, they can be used as chemical tools to study other natural products. In an example of this application, the Balskus lab used one inhibitor to block the activation of two toxins in the native organisms and determine the chemical structure of the toxin precursors. Thus, the new inhibitors are already proving that they will be powerful tools both for studying the impacts of colibactin on host health, and for investigating natural product biosynthesis more broadly.
by José A. Velilla and Rachelle Gaudet
Velilla et al.:
Volpe et al.:



