Many functional RNAs such as tRNAs or rRNA adopt 3D structures that are specific to their distinct functions and conserved across species. As with proteins, the determination of RNA structures relies on experimental and costly methods such as x-Ray crystallography, NMR, and cryo-EM. Motivated by proteins, currently there is a race to a “AlphaFold moment“ for RNA. Highly accurate machine learning (ML) predictions of RNA structures would allow us to make fast and reliable functional hypotheses of molecular mechanisms involving RNA without the need to wait for a crystal structure.
The determination of RNA structure by ML requires training and testing sets of reliable RNA 3D structures. Those are usually obtained from the Protein Data Bank (PDB), the repository by excellence of all known experimentally-determined protein and structural RNAs. Importantly, ML methods “learn” from the training set, and use the representation obtained from the training structures to predict the structures in the test sets. To determine whether a method has actually learned the rules of RNA structure, or it has simply memorized the sequences and, importantly, the structures present in the training set, ML-methods need to use testing sets that are structurally dissimilar from those in the training set. This somewhat obvious train/test split requirement, that we identified for early ML methods of RNA structure prediction (Rivas et al., 2012), has been somewhat overlooked by recent deep-learning methods (Szikszai et al., 2022).
In this Journal of Molecular Biology (PDF) article, Elena Rivas with Marcell Szikszai and Marcin Magnus in her group (and collaborators from the OpenFold team, a trainable reimplementation of AlphaFold) introduce the method RNA3DB. RNA3DB partitions the PDB RNA structures into training and testing sets that can be reliably used by any ML-RNA structure prediction method to investigate their generalization potential.
Many methods before have explored the complexities of PDB with respect to RNA structure, and many databases classifying RNA PDB structures exist. In this work, RNA3DB sets itself to achieve one unique, important and yet unresolved goal, that of producing a structurally dissimilar split of the RNA structures on PDB for training and testing ML models of RNA structure.
RNA3DB uses the RNA homology method Infernal, the database of structural RNAs Rfam, and a graph-theorical method to approach with rigor the question of ML generalization by building rigorous training and testing databases of PDB RNA structures guaranteed to be structurally dissimilar from each other. The RNA3DB method is highly customizable, and the RNA3DB dataset is updated regularly.
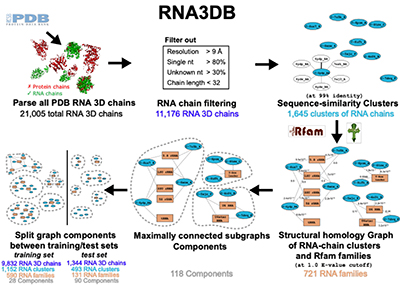
With the RNA3DB methodology in hand, we can now concentrate on other subsequent and important questions: Do we have enough RNA structural data to be able to build an AlphaFold for RNA? As it stands, while AlphaFold used several hundreds of thousand different PDB protein structures alone to train its models, there is only of the order of ten thousand total RNA structures in PDB, of which less than 2,000 correspond to unique sequences (See Figure).
 (PDF)
(PDF)


